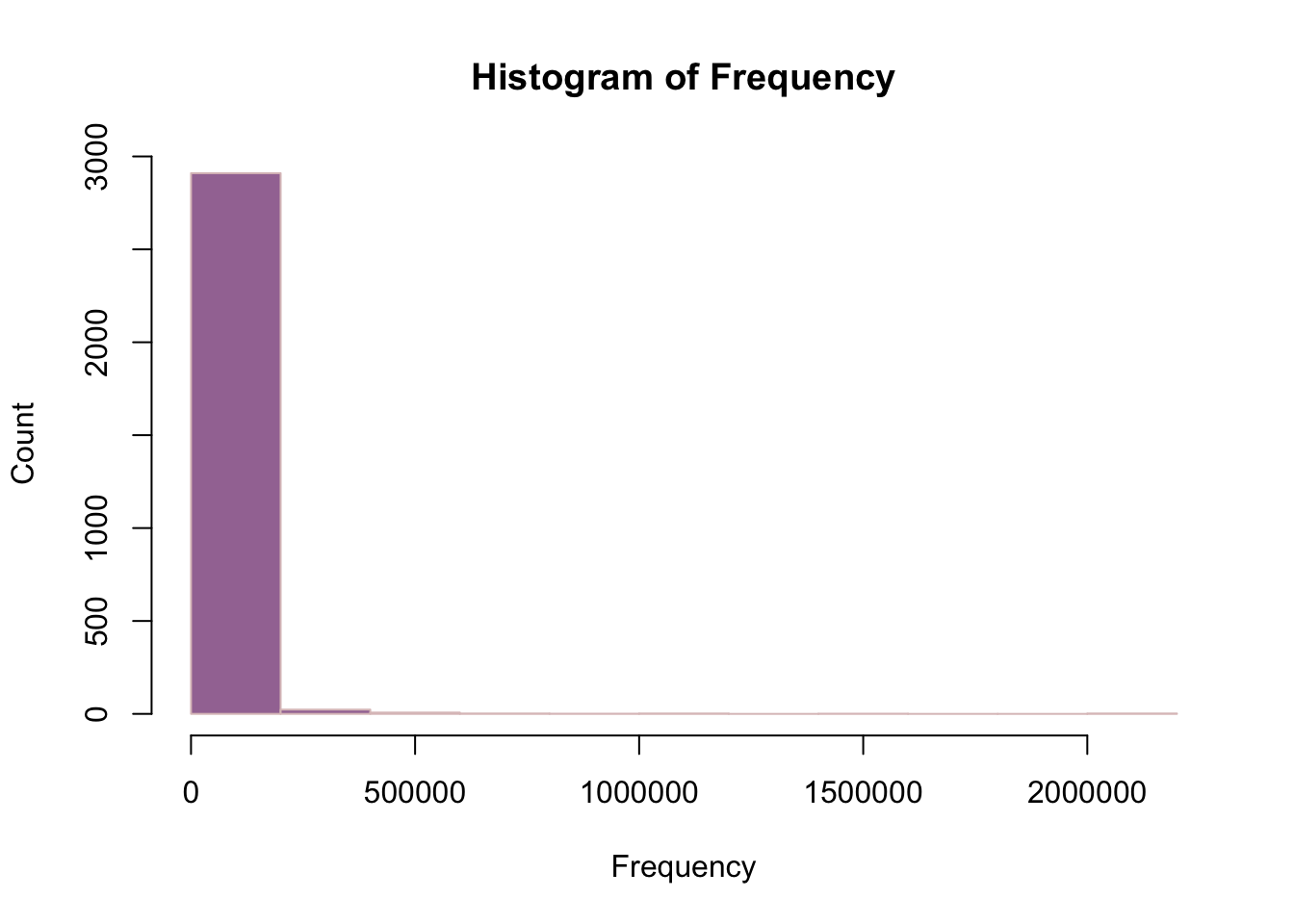
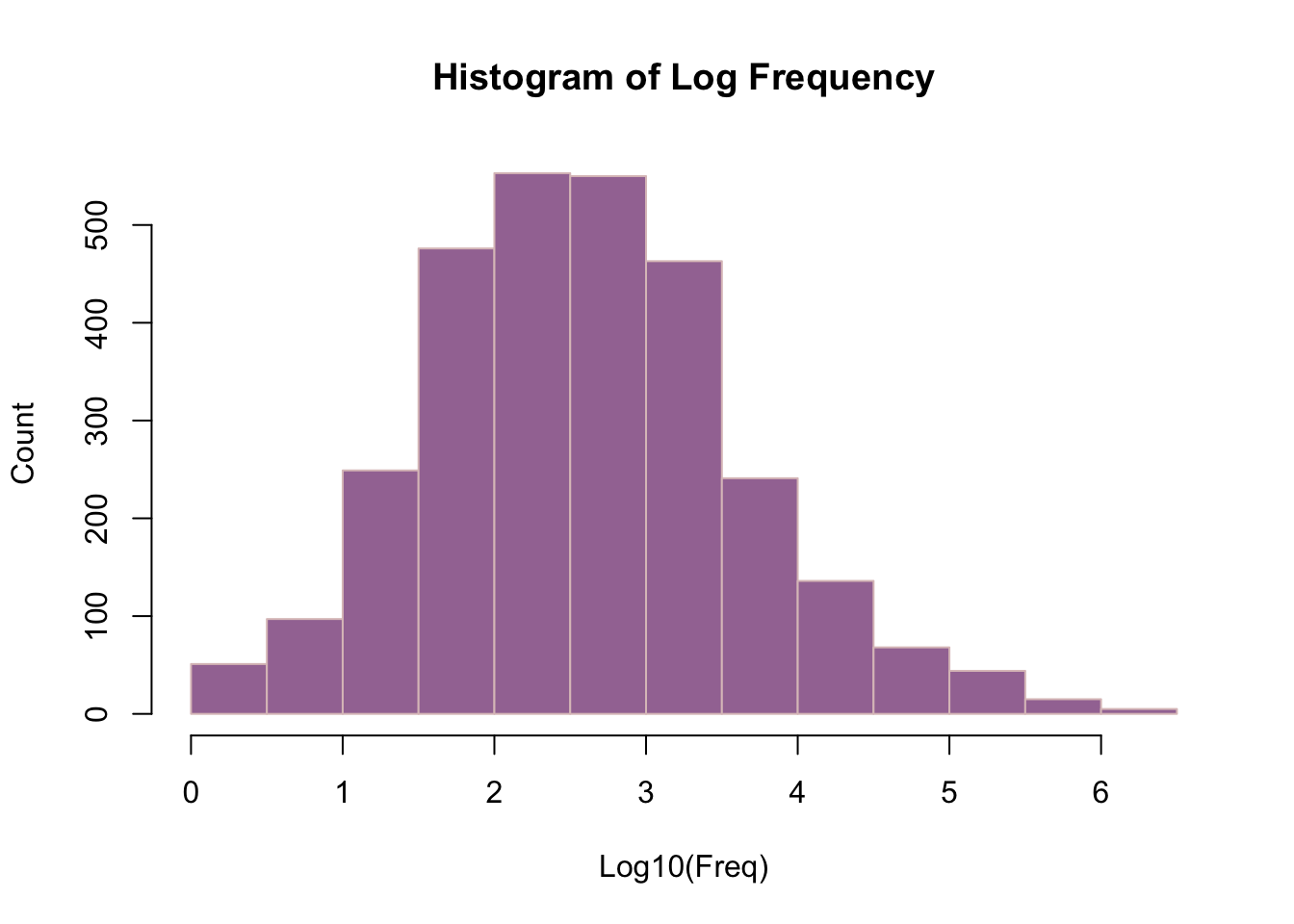
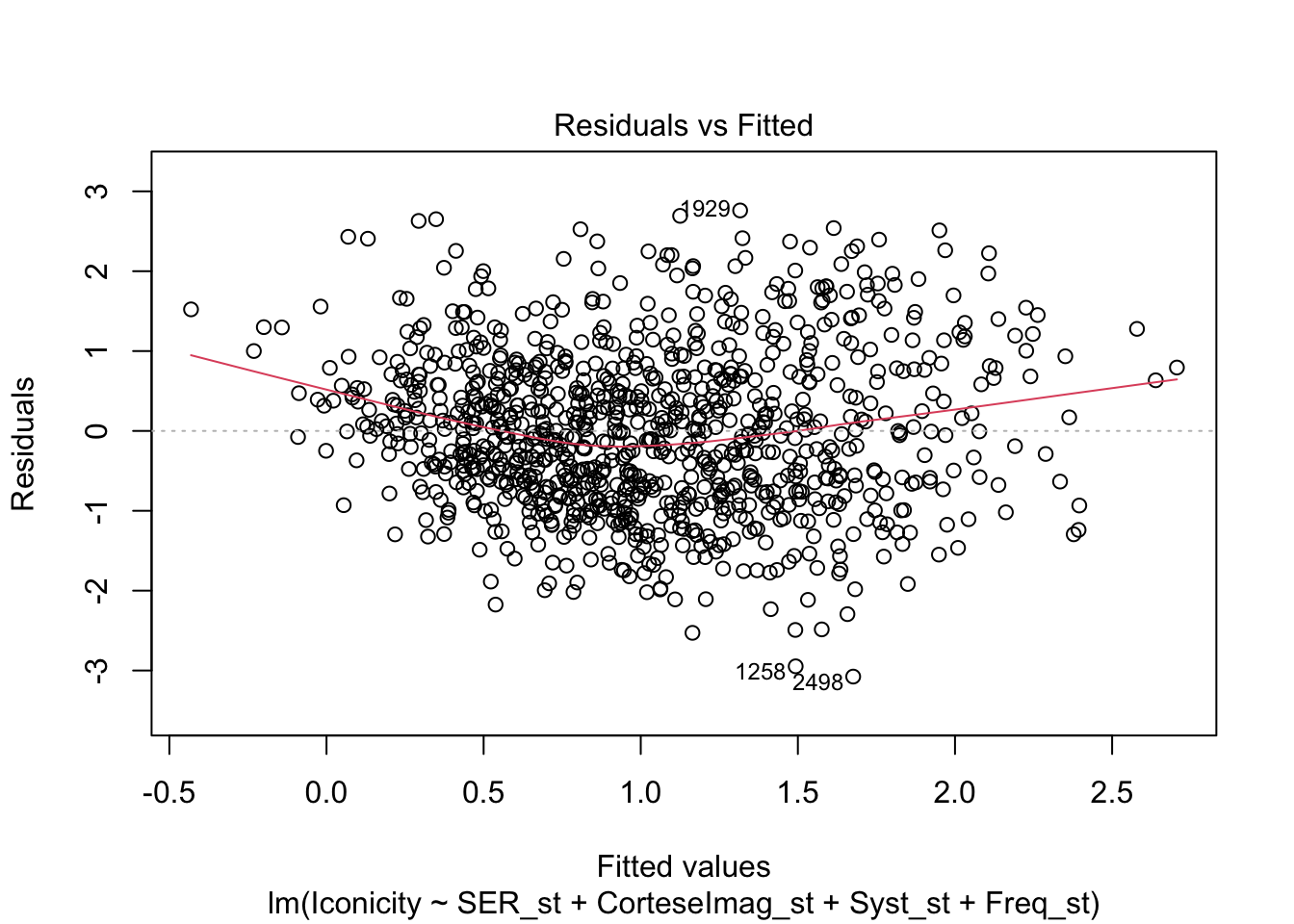
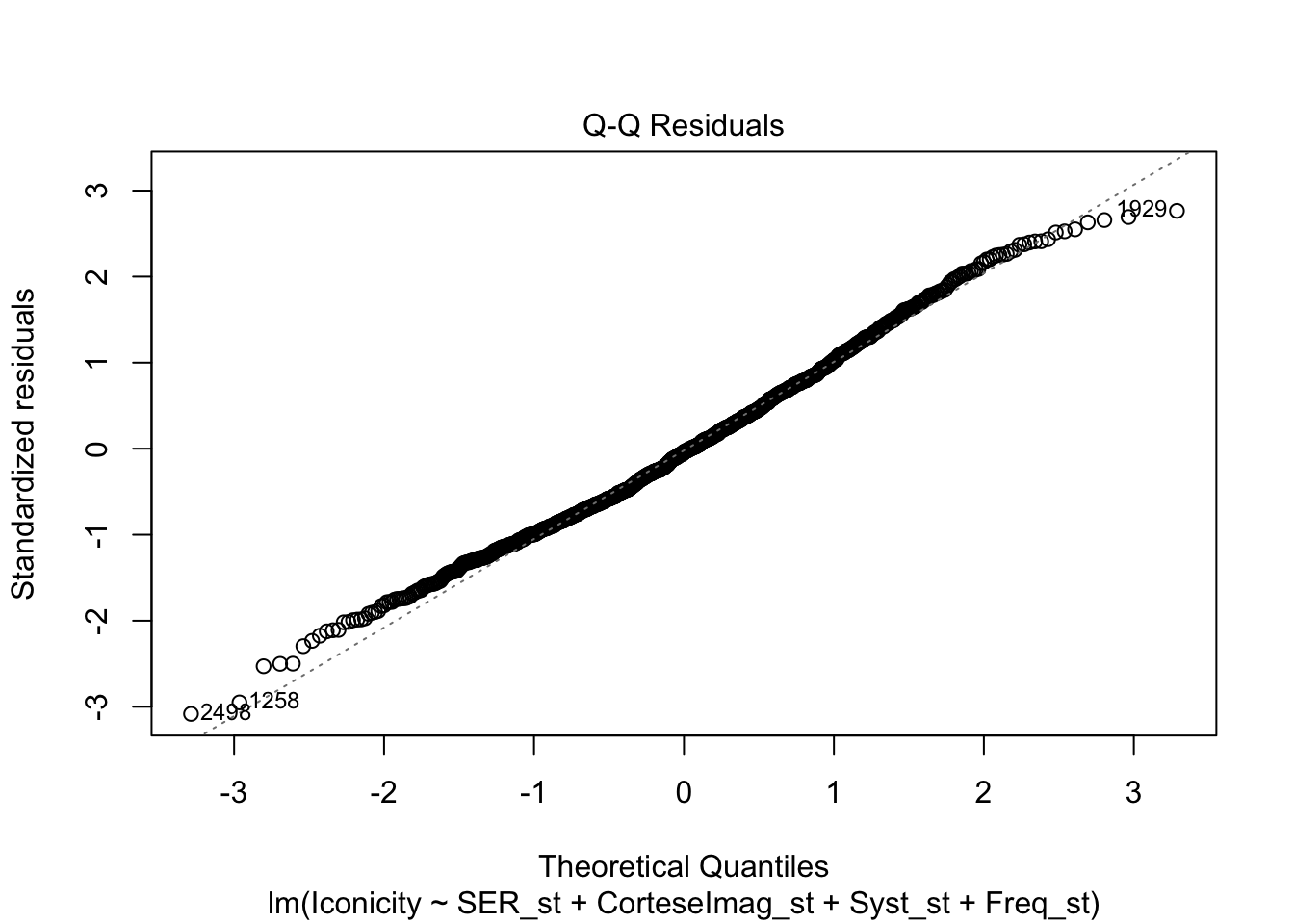
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.1 ✔ stringr 1.5.2
✔ ggplot2 4.0.0 ✔ tibble 3.3.0
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.1.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorslibrary(broom)
rawdata<- read_csv("winter_2017_iconicity.csv")Rows: 3001 Columns: 8
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (2): Word, POS
dbl (6): SER, CorteseImag, Conc, Syst, Freq, Iconicity
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.rawdata %>% print(n=10, width=Inf) # width = Inf is to see all the columns of the tibble# A tibble: 3,001 × 8
Word POS SER CorteseImag Conc Syst Freq Iconicity
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 a Grammatical NA NA 1.46 NA 1041179 0.462
2 abide Verb NA NA 1.68 NA 138 0.25
3 able Adjective 1.73 NA 2.38 NA 8155 0.467
4 about Grammatical 1.2 NA 1.77 NA 185206 -0.1
5 above Grammatical 2.91 NA 3.33 NA 2493 1.06
6 abrasive Adjective NA NA 3.03 NA 23 1.31
7 absorbent Adjective NA NA 3.1 NA 8 0.923
8 academy Noun NA NA 4.29 NA 633 0.692
9 accident Noun NA NA 3.26 NA 4146 1.36
10 accordion Noun NA NA 4.86 NA 67 -0.455
# ℹ 2,991 more rows